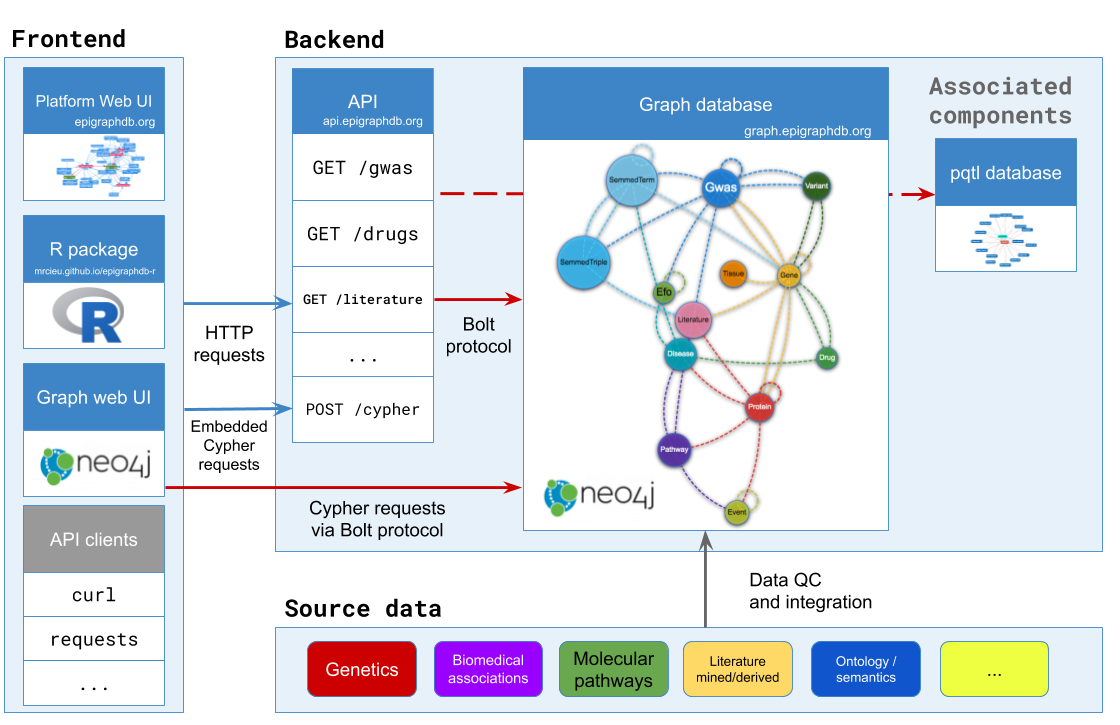
About EpiGraphDB
The increasing availability of complex, high-dimensional epidemiological data necessitates innovative and scalable approaches to harness its potential to address research questions of biomedical importance. EpiGraphDB is an analytical platform and database that aims to address this challenge, supporting data mining in epidemiology.
Our core objectives are to:
- Develop approaches for the appropriate application and interpretation of causal inference in systematic automated analyses of many phenotypes using data from a rich array of bioinformatic resources.
- Apply data mining approaches to the same integrated dataset to make novel discoveries about disease mechanisms and potential interventions relevant to population health.
EpiGraphDB is developed by members of the data mining programme at the MRC Integrative Epidemiology Unit.
EpiGraphDB resources
Web application
Visit https://epigraphdb.org .
Web API
Visit https://api.epigraphdb.org .
R package
Funding sources
- EpiGraphDB receives core funding from the UK Medical Research Council as part of the Data Mining Epidemiological Relationships programme in the MRC Integrative Epidemiology Unit.
- The pQTL browser was developed as part of a collaboration between the MRC IEU, GlaxoSmithKline and Biogen, and is described here.
- The MR-EvE data within EpiGraphDB has been produced by Gibran Hemani on a Wellcome Sir Henry Dale fellowship.
- Pathway data and network analysis methods have been supported by funding from Cancer Research UK.
Key technologies
Below are some of the key technologies that power EpiGraphDB:
Citation
Please cite EpiGraphDB as
Yi Liu, Benjamin Elsworth, Pau Erola, Valeriia Haberland, Gibran Hemani, Matt Lyon, Jie Zheng, Oliver Lloyd, Marina Vabistsevits, Tom R Gaunt, EpiGraphDB: a database and data mining platform for health data science, Bioinformatics, btaa961, https://doi.org/10.1093/bioinformatics/btaa961
@article{epigraphdb2020bioinformatics,
author = {Liu, Yi and Elsworth, Benjamin and Erola, Pau and Haberland, Valeriia and Hemani, Gibran and Lyon, Matt and Zheng, Jie and Lloyd, Oliver and Vabistsevits, Marina and Gaunt, Tom R},
title = {{EpiGraphDB}: a database and data mining platform for health data science},
journal = {Bioinformatics},
year = {2020},
month = {11},
issn = {1367-4803},
doi = {10.1093/bioinformatics/btaa961},
url = {https://doi.org/10.1093/bioinformatics/btaa961},
note = {btaa961},
eprint = {https://academic.oup.com/bioinformatics/advance-article-pdf/doi/10.1093/bioinformatics/btaa961/34178613/btaa961.pdf}
}
Contact
Please get in touch with us for issues, comments, suggestions, etc. via the following methods: