EpiGraphDB graph database
The EpiGraphDB database is built using neo4j, a graph database technology:
Below is a simple representation of EpiGraphDB as a meta graph (http://epigraphdb.org/about):

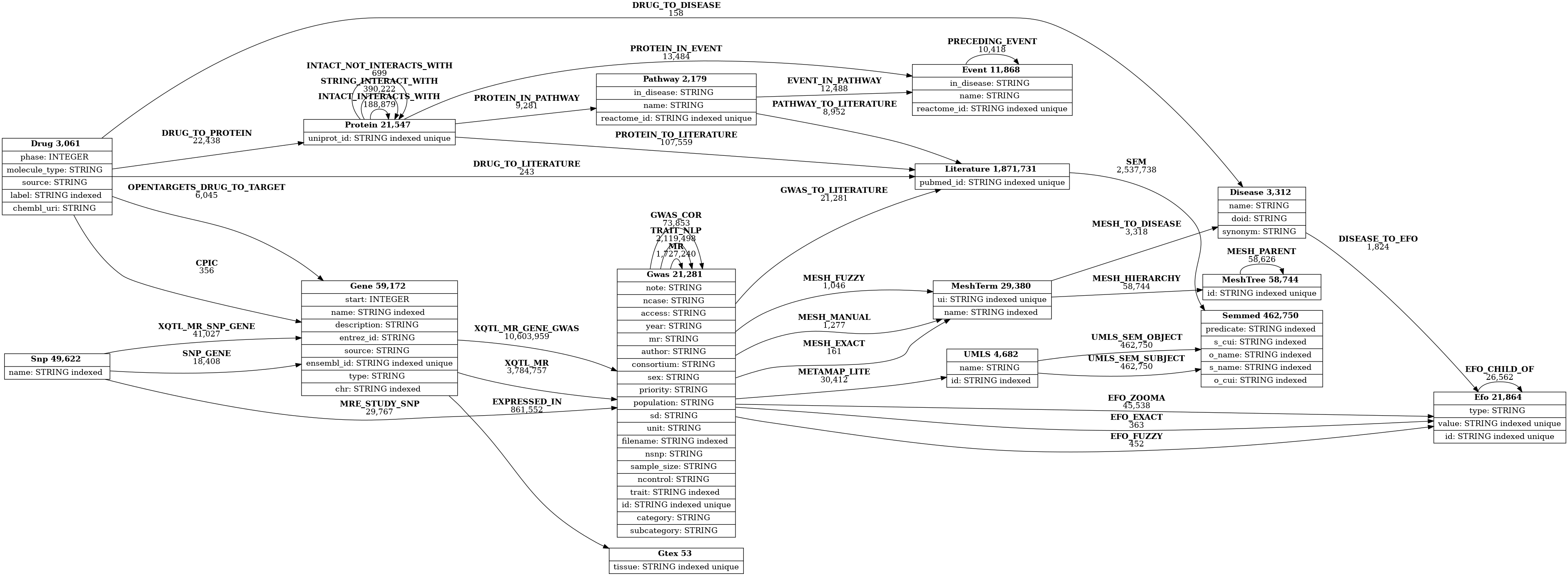
And below is the schema of the graph database (http://api.epigraphdb.org/meta/schema?graphviz=true&plot=true):
Data sources
EpiGraphDB integrates data generated at the MRC IEU with data from a range of third party sources. These include1:
- UK Biobank genome-wide association study results from the IEU GWAS database: https://gwas.mrcieu.ac.uk
- MR-EvE causal estimates: https://doi.org/10.1101/173682
- Reactome pathway data: https://reactome.org/
- STRING protein association data: https://string-db.org/
- IntAct molecular interaction data: https://www.ebi.ac.uk/intact/
- Semantic predications from SemMedDB: https://skr3.nlm.nih.gov/SemMedDB/
- Experimental factor ontology (EFO) terms: https://www.ebi.ac.uk/efo/
- Drug-target relationships from Open Targets: https://www.targetvalidation.org/
- Gene-drug relationships from Clinical Pharmacogenetics Implementation Consortium (CPIC): https://cpicpgx.org/genes-drugs/
- Gene and Protein data from BioMart Ensembl: http://www.ensembl.org/biomart
- Tissue specific gene expression data from GTEx: https://gtexportal.org/home/
- Disease data from Mondo Disease Ontology: https://mondo.monarchinitiative.org/
- Druggable genes: https://stm.sciencemag.org/content/suppl/2017/03/27/9.383.eaag1166.DC1
- Expression quantitative trait locus (eQTL) data from the eQTLGen consortium: https://www.eqtlgen.org/
- Protein quantitative trait locus (pQTL) data from plasma proteins: http://epigraphdb.org/risk-factor-drugs/
- GWAS to UMLS relationships: https://metamap.nlm.nih.gov/MetaMapLite.shtml
- Polygenic Risk Score from PRS Atlas http://mrcieu.mrsoftware.org/PRS_atlas/
- Genetic correlation data from Neale Lab https://ukbb-rg.hail.is/
- Observational correlation data from UK Biobank https://www.ukbiobank.ac.uk/
- xQTL study project Zheng et al., forthcoming
- Vectology http://vectology.mrcieu.ac.uk
- Monarch Initiative https://monarchinitiative.org/
-
For the full list of source data and how they are integrated into EpiGraphDB please refer to our documentation on meta nodes and meta relationships. ↩